New UMHET3 and aging portal! This is initial work for providing a full community webservice for aging.
- https://doi.org/10.1101/2025.04.27.649857. Genetic Modulation of Lifespan: Dynamic Effects, Sex Differences, and Body Weight Trade-offs
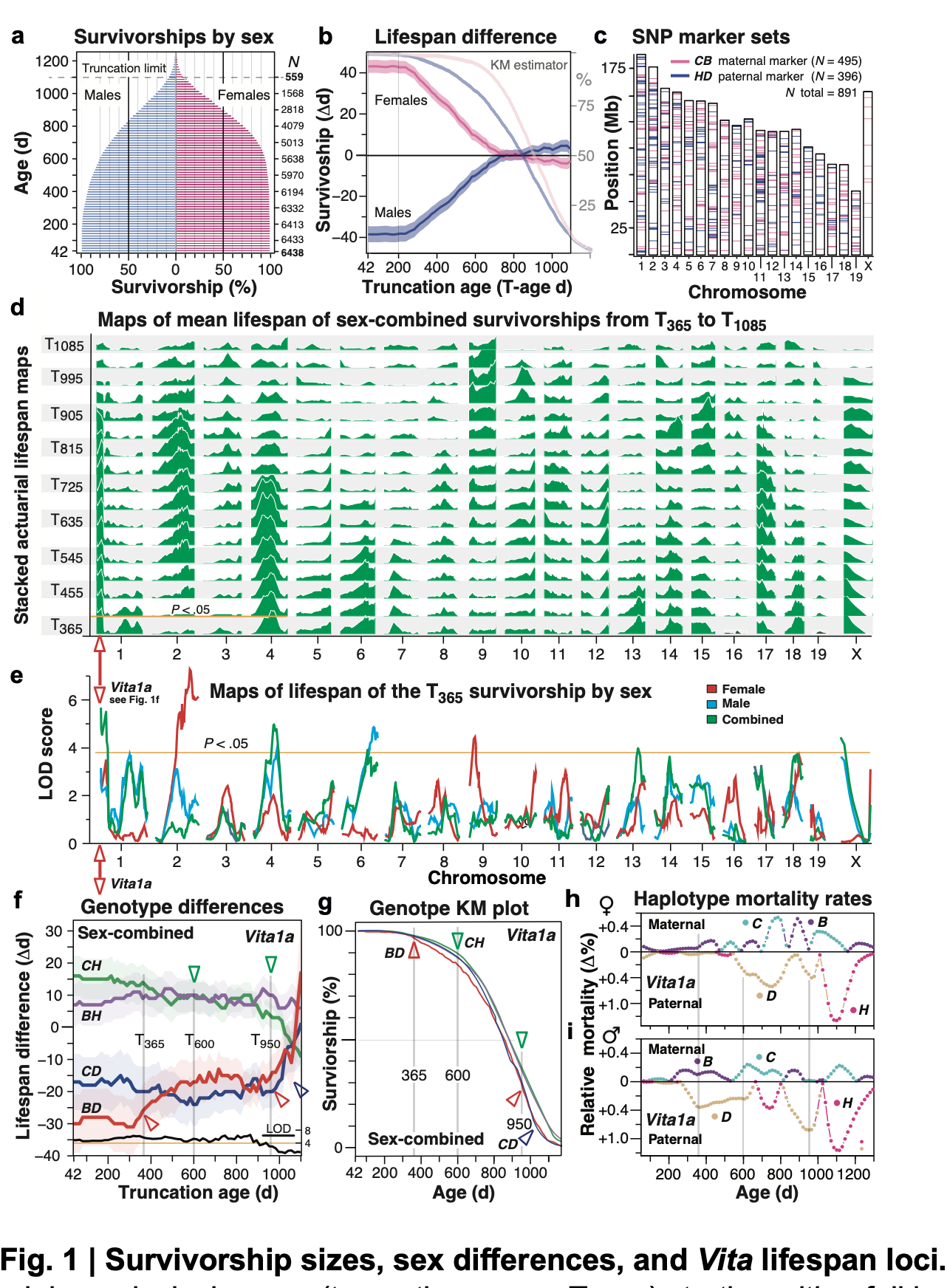
Abstract: The dynamics of lifespan are shaped by DNA variants that exert effects at different ages. We have mapped genetic loci that modulate age-specific mortality using an actuarial approach. We started with an initial population of 6,438 pubescent siblings and ended with a survivorship of 559 mice that lived to at least 1100 days. Twenty-nine Vita loci dynamically modulate the mean lifespan of survivorships with strong age- and sex-specific effects. Fourteen have relatively steady effects on mortality while other loci act forcefully only early or late in life and with polarities of effects that invert. A distinct set of 19 Soma loci shape the negative correlation between weights of young adults with their life expectancies—much more strongly so in males than females. Another set of 11 Soma loci shape the positive correlation between weights at older ages with life expectancies. The Vita and Soma loci share 289 age-dependent epistatic interactions (LODs ≥3.8) but fewer than 4% are common to both sexes. More....
We provide two examples of how to move from maps toward potential mechanisms. Our findings provide an empirical bridge between evolutionary theories on aging and genetic and molecular causes. These loci and their interactions are key to begin to understand the impact of interventions that may extend healthy lifespan in mice and even in humans.
- Data files are available here.
- Code is available at https://github.com/DannyArends/UM-HET3/
See also GeneNetwork.org examples on aging.
Explore GeneNetwork2 for aging (GN4AGING)
Find all genes that have a hit for
Find all phenotypes that have a hit for
Note that you can use the powerful search at the top of the results page! Search terms can be added. In the results click on any item to (re)run the relevant GWA or QTL mapping
Contact
We host private and public datasets. If you are interested in exploring data on GeneNetwork, or to add your own data to benefit from the power of integrated datasets, please contact:
David Ashbrook | Rob W Williams | Pjotr Prins